[!+] Plot a confusion matrix (a.k.a. classification table)
qplot_confusion.RdPlot a confusion matrix (classification table) with
additional statistics (sensitivity (Se) a.k.a. true positive
rate, positive predictive value (PPV) and Cohens' Kappa (k)).
Colors in the main matrix: diagonal cells vary from grey to
green and offdiagonal elements vary from grey to
red. The color intensity in certain cell is determined by
parameter shades.
Colors of cells with statistics Se, PPV and k vary depening on
values of these statistics from red (low values) to
grey (middle values), to green (high values).
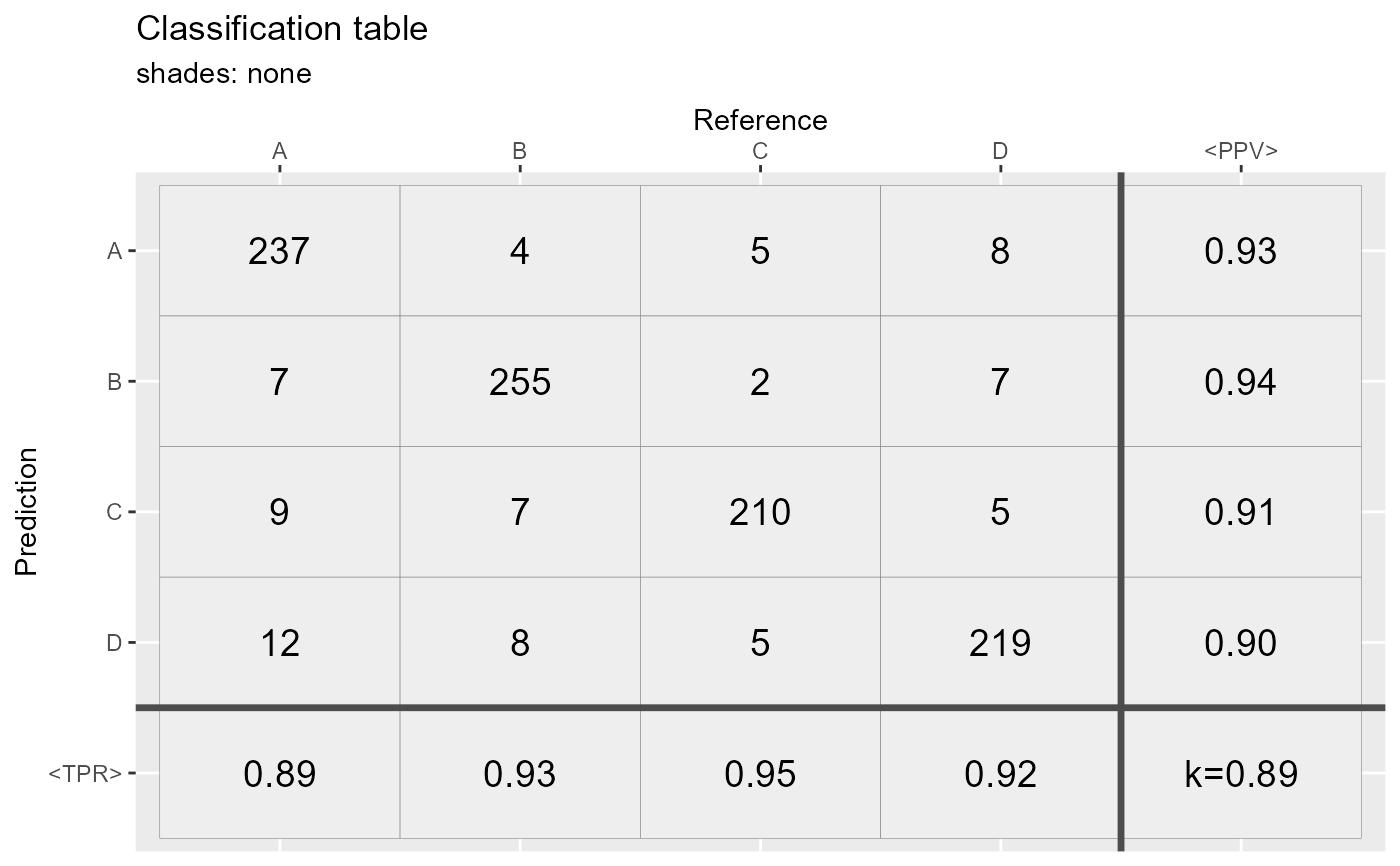
Exception: when shades="const" and shades="none",
color intensities are constant.
qplot_confusion(...)
# S3 method for default
qplot_confusion(
Prediction,
Reference,
Title = "Classification table",
xLabel = NULL,
yLabel = NULL,
subTitle = NULL,
shades = c("prop", "max", "const", "none"),
guide = FALSE,
text.size = 5,
decimals = 2,
...
)
# S3 method for matrix
qplot_confusion(mat, ...)
# S3 method for ResampleResult
qplot_confusion(obj, ...)
# S3 method for PredictionClassif
qplot_confusion(obj, ...)
# S3 method for table
qplot_confusion(
conf,
Title = "Classification table",
xLabel = NULL,
yLabel = NULL,
subTitle = NULL,
shades = c("prop", "max", "const", "none"),
guide = FALSE,
text.size = 5,
decimals = 2,
TPR.name = "<TPR>",
PPV.name = "<PPV>",
sort = c(FALSE, "diagonal", "PPV", "TPR"),
metric = c("kappa", "weighted.kappa", "meanTPR", "meanPPV")
)Arguments
- ...
Appropriate parameters (described below).
- Prediction
A factor variable with predicted groups.
- Reference
A factor variable with reference groups.
- Title
The main title for the plot.
- xLabel
A label for abscisa (x) axis.
- yLabel
A label for ordinate (y) axis.
- subTitle
The second line of title, which will be smaller and and in italics.
- shades
A function how intensities of cell colors in the main confsion matrix depend on cell values:
- "prop"
An intensity of a cell color is proportianal to a value in the cell. Distribution of color intensities is balanced according to number of classes (but not balanced according to number of observations per class.)
- "max"
Cell with the absolute maximum value is represented by the most intensive color. Other cells are proportionally less intensive.
- "const"
Constant red and green colors.
- "none"
All cells are grey.
- guide
Logical. If
TRUE, a legend is plotted.- text.size
The size of a text inside cells.
- decimals
The number of decimal positions in rounding. Default is 2 (i.e., precission is 0.01).
- conf, mat
A confusion matrix (classification table): either an object of a class "table" or a square matrix.
- TPR.name
The name of the row with true positive rate (TPR). Default is "<TPR>". It is the same as "<Sensitivity>".
- PPV.name
The name of the column with positive predictive value (PPV). Default is "<PPV>".
- sort
A way to sort columns and rows of output matrix. Options to sort values in descending order:
FALSE(default) do not sort;"diagonal"- sort values on diagonal;"PPV"- sort positive predictive values (PPV);"TPR"- sort true positive rate (TPR) values.- metric
The metric of overall accuracy. Curently supported values are:
"kappa"(default) to use Cohen's kappa, in table denoted with symbol "k";"weighted.kappa"to use weighted kappa, (symbol "w");"meanTPR"Mean of true positive rates of each group (symbol "t");"meanPPV"Mean of positive predictive values of each group (symbol "v").
Value
A plot of confusion matrix and additional statistics (`ggplot` object).
See also
Other spHelper plots:
check_palette(),
layer_spRangeMean(),
plot_colors(),
plot_hyPalette(),
plot_spCompare(),
plot_spDiff(),
plot_spDistribution(),
qplot_crosstab(),
qplot_infoDim(),
qplot_kAmp(),
qplot_kSp(),
qplot_prediction(),
qplot_spRangeCenter(),
qplot_spRangeMedian(),
qplot_spStat(),
qplot_spc(),
rmExpr(),
rm_stripes(),
stat_chull()
Examples
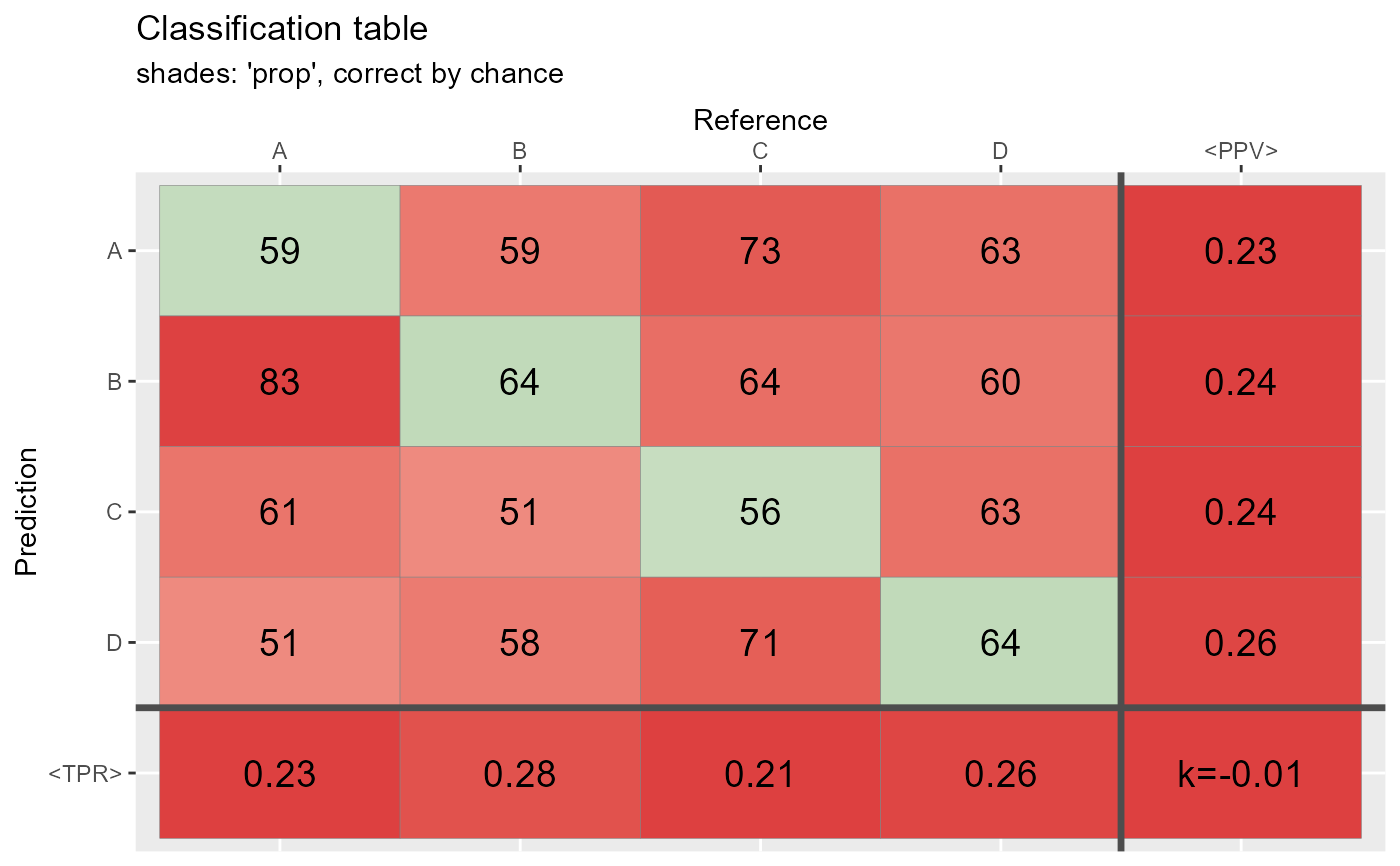
# Generate data: Random guess ============================
N <- 1000 # number of observations
Prediction <- sample(c("A","B","C","D"), N, replace = TRUE)
Reference <- sample(c("A", "B","C","D"),N, replace = TRUE)
# This function:
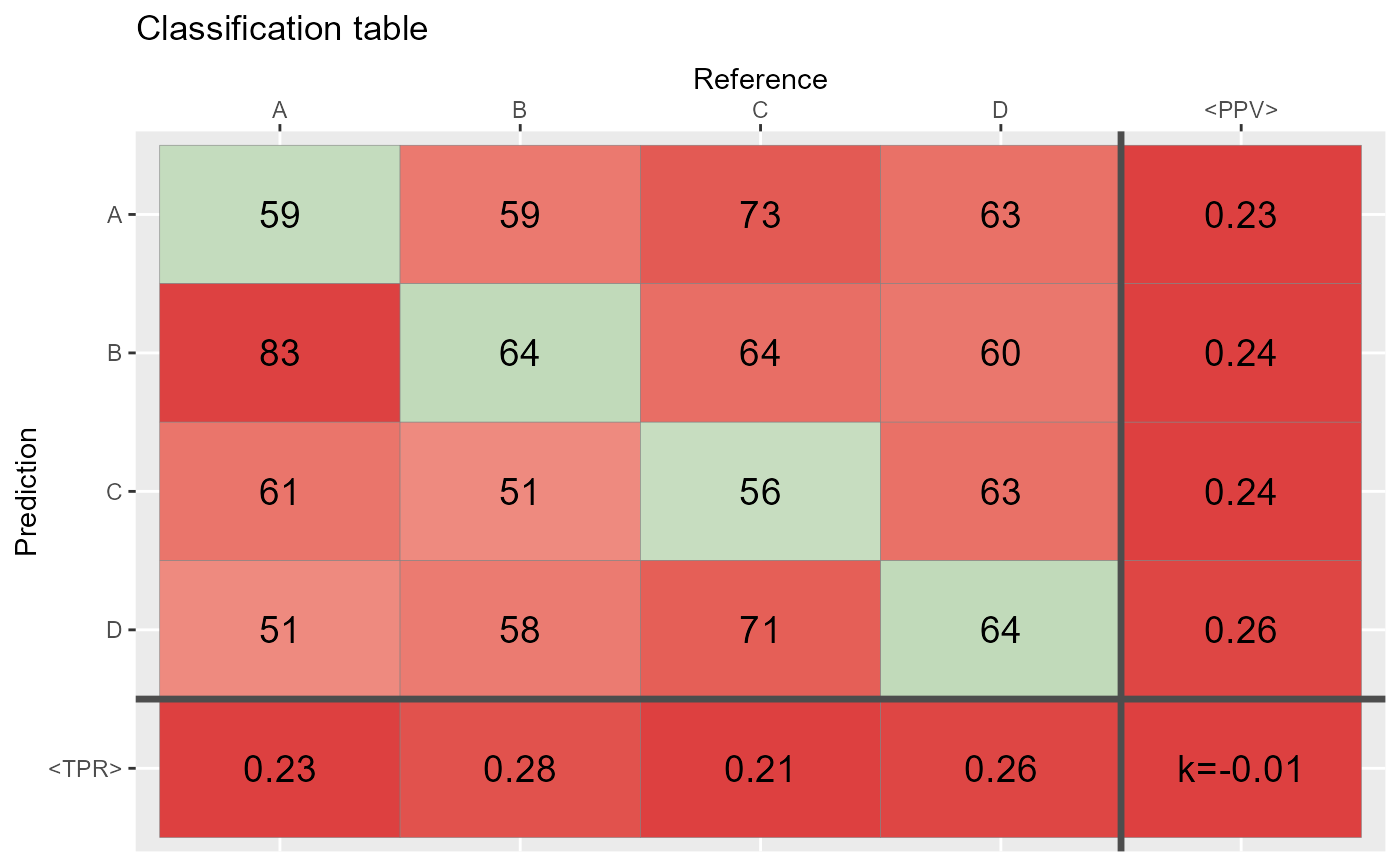
qplot_confusion(Prediction, Reference)
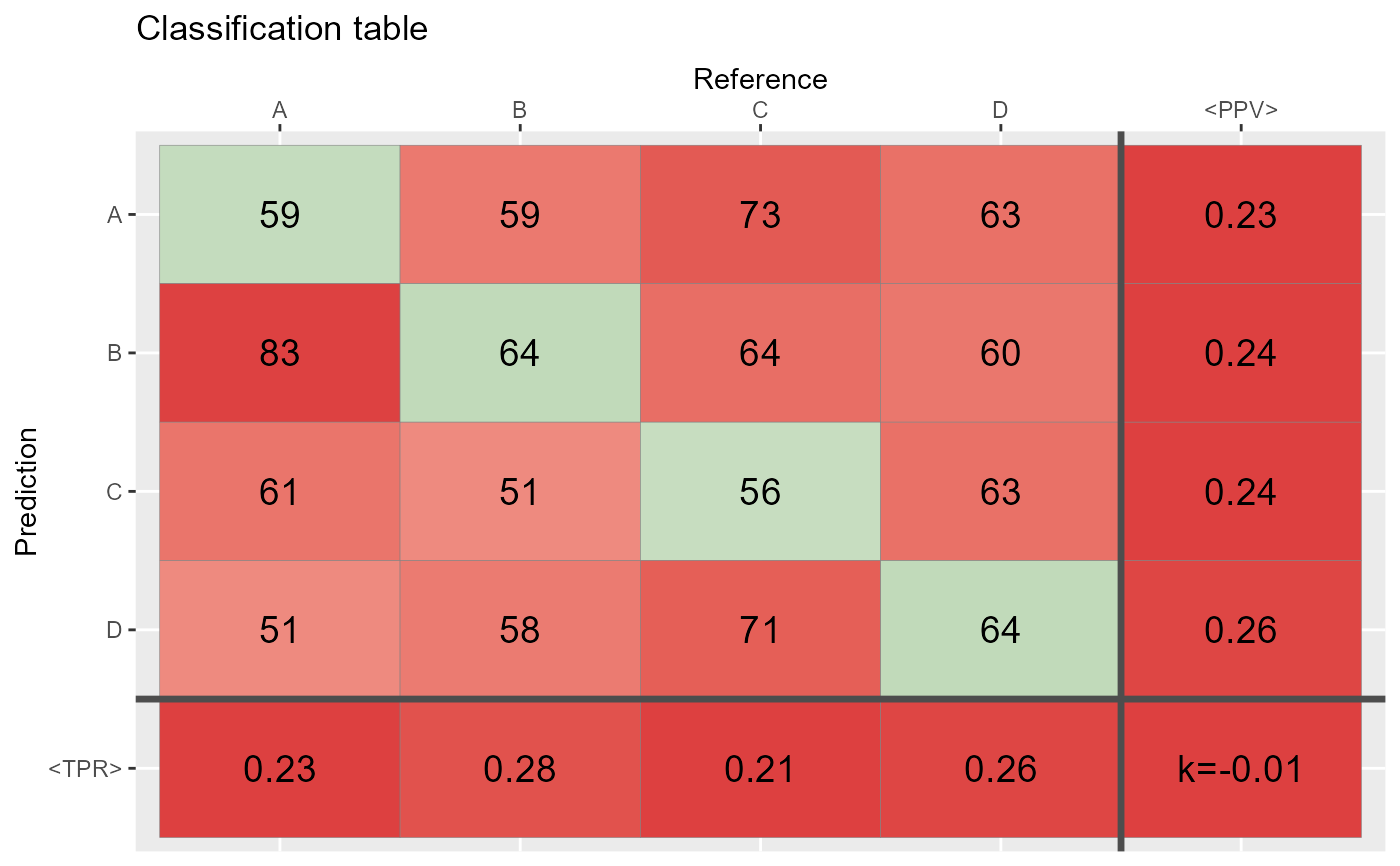 # does the same as:
conf <- table(Prediction,Reference)
qplot_confusion(conf)
# does the same as:
conf <- table(Prediction,Reference)
qplot_confusion(conf)
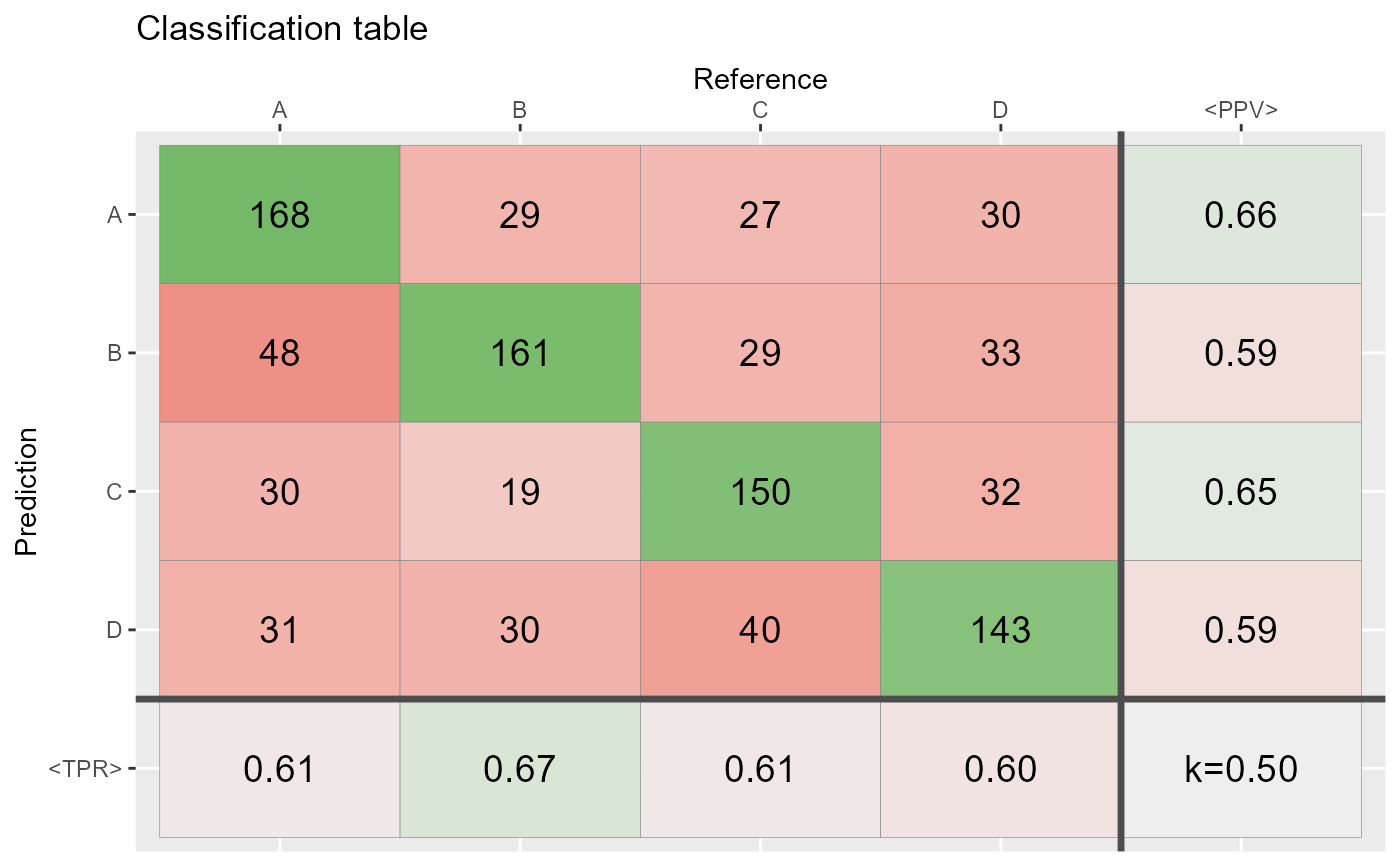
 # At least 50% of the cases agree =========================
ind <- sample(1:N,round(0.50*N))
Reference[ind] <- Prediction[ind]
conf2 <- table(Prediction,Reference)
qplot_confusion(conf2)
# At least 50% of the cases agree =========================
ind <- sample(1:N,round(0.50*N))
Reference[ind] <- Prediction[ind]
conf2 <- table(Prediction,Reference)
qplot_confusion(conf2)
 # Most of the cases agree =================================
ind <- sample(1:N,round(N*.80))
Reference[ind] <- Prediction[ind]
conf3 <- table(Prediction,Reference)
qplot_confusion(conf3)
# Most of the cases agree =================================
ind <- sample(1:N,round(N*.80))
Reference[ind] <- Prediction[ind]
conf3 <- table(Prediction,Reference)
qplot_confusion(conf3)
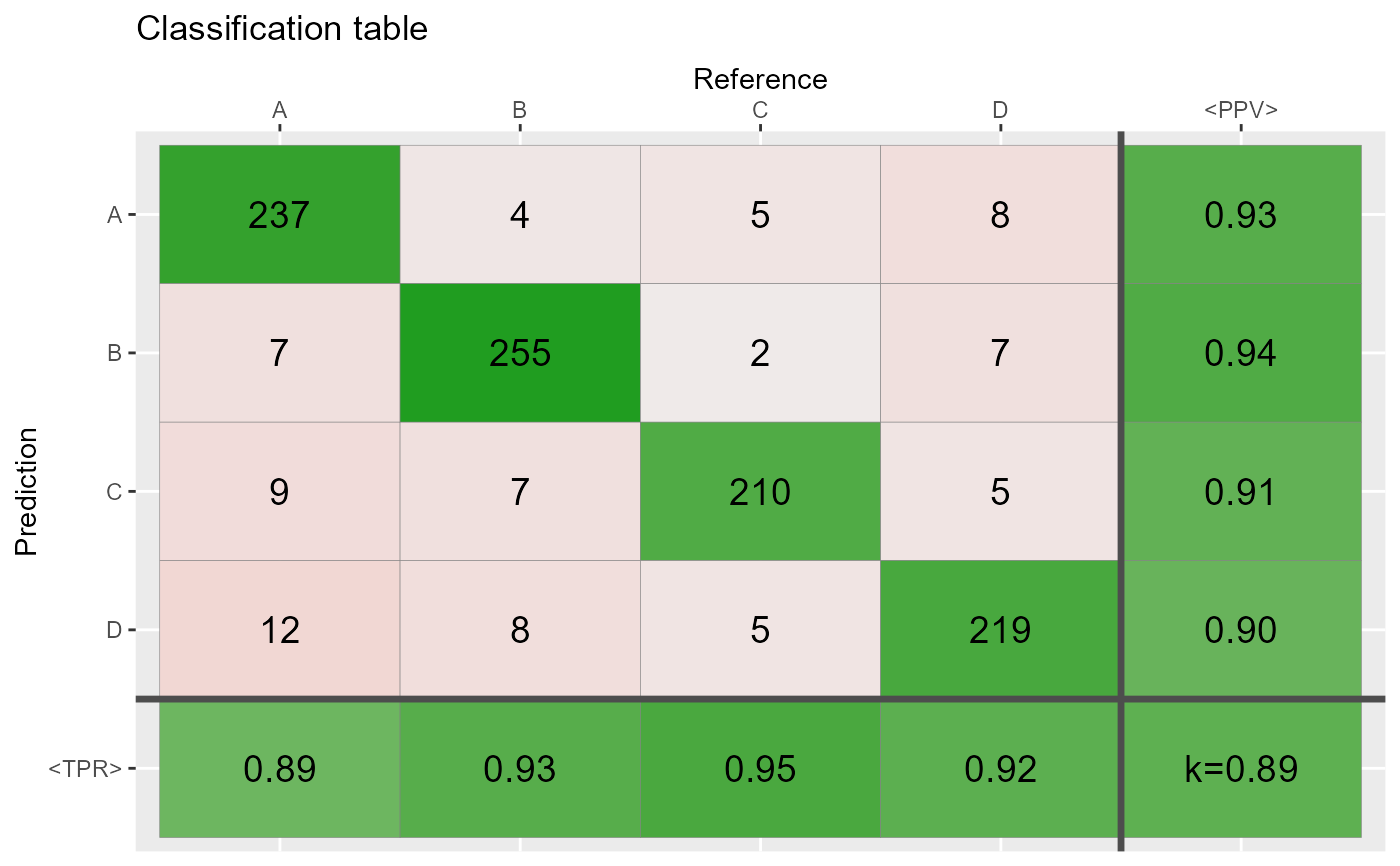 # Proportions =============================================
qplot_confusion(conf3)
# Proportions =============================================
qplot_confusion(conf3)
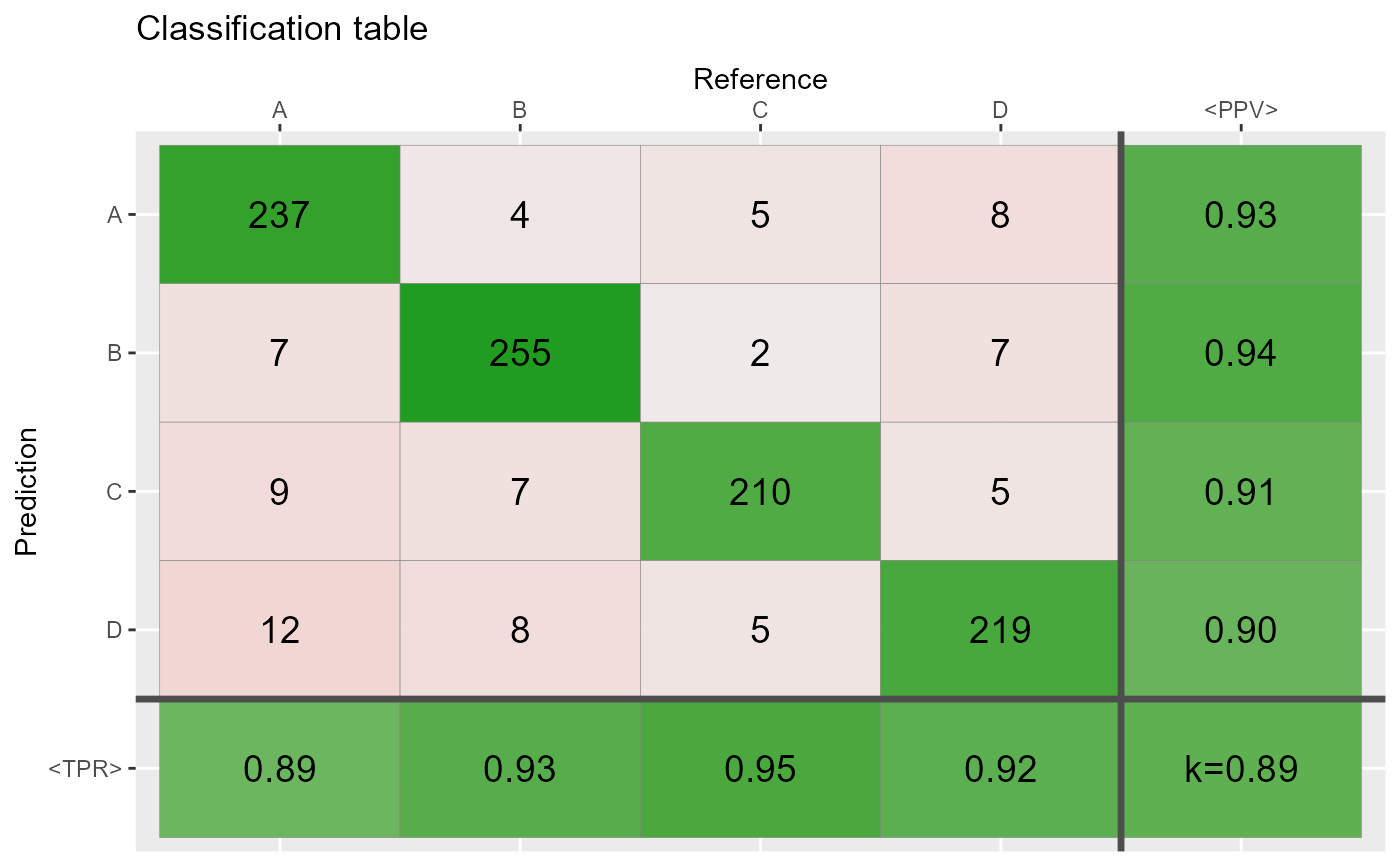 # Shades: proportional =====================================
qplot_confusion(conf,shades = "prop", subTitle = "shades: 'prop', correct by chance")
# Shades: proportional =====================================
qplot_confusion(conf,shades = "prop", subTitle = "shades: 'prop', correct by chance")
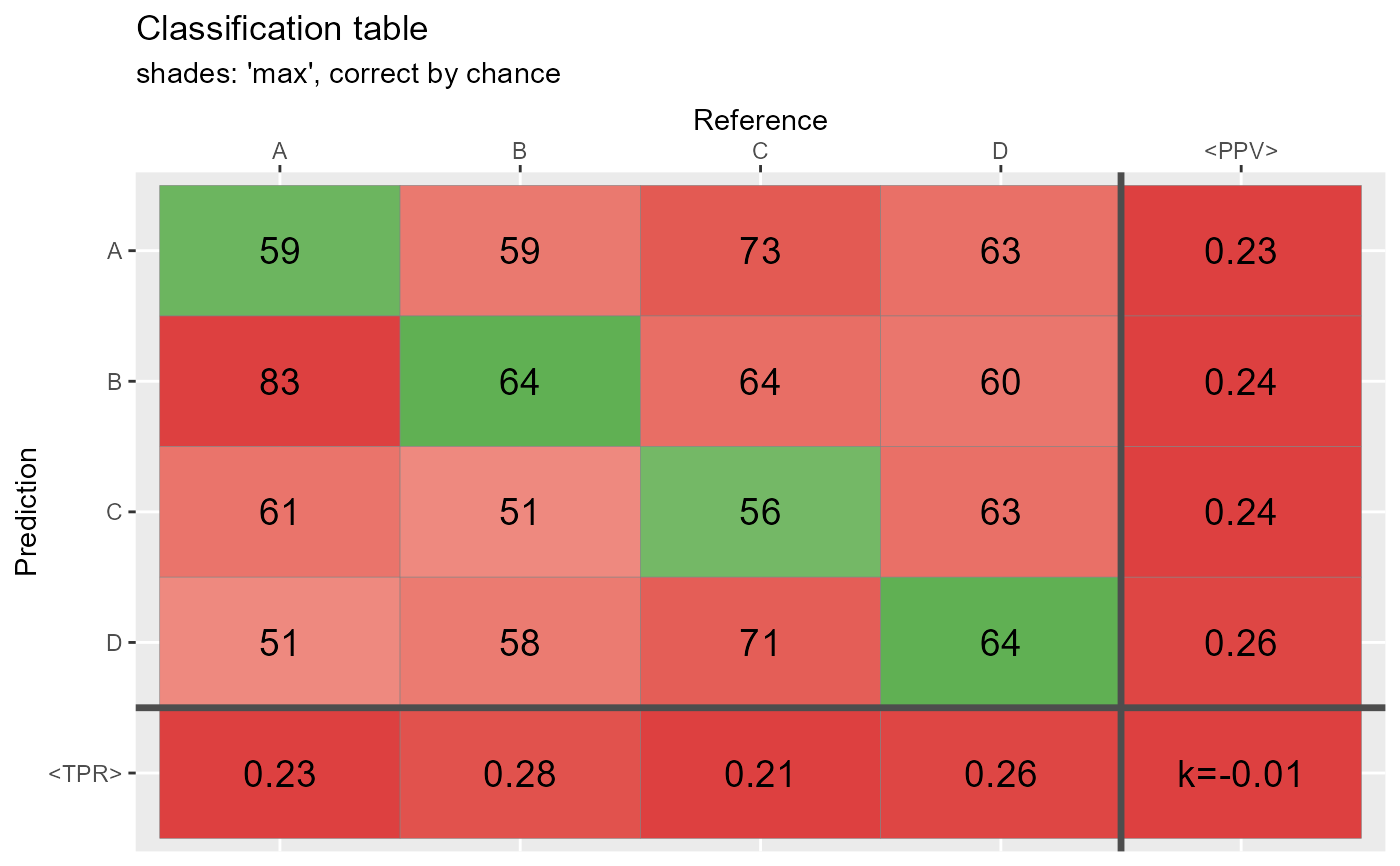
 qplot_confusion(conf,shades = "max", subTitle = "shades: 'max', correct by chance")
qplot_confusion(conf,shades = "max", subTitle = "shades: 'max', correct by chance")
 qplot_confusion(conf2,shades = "prop", subTitle = "shades: 'prop', correct >50%")
qplot_confusion(conf2,shades = "prop", subTitle = "shades: 'prop', correct >50%")
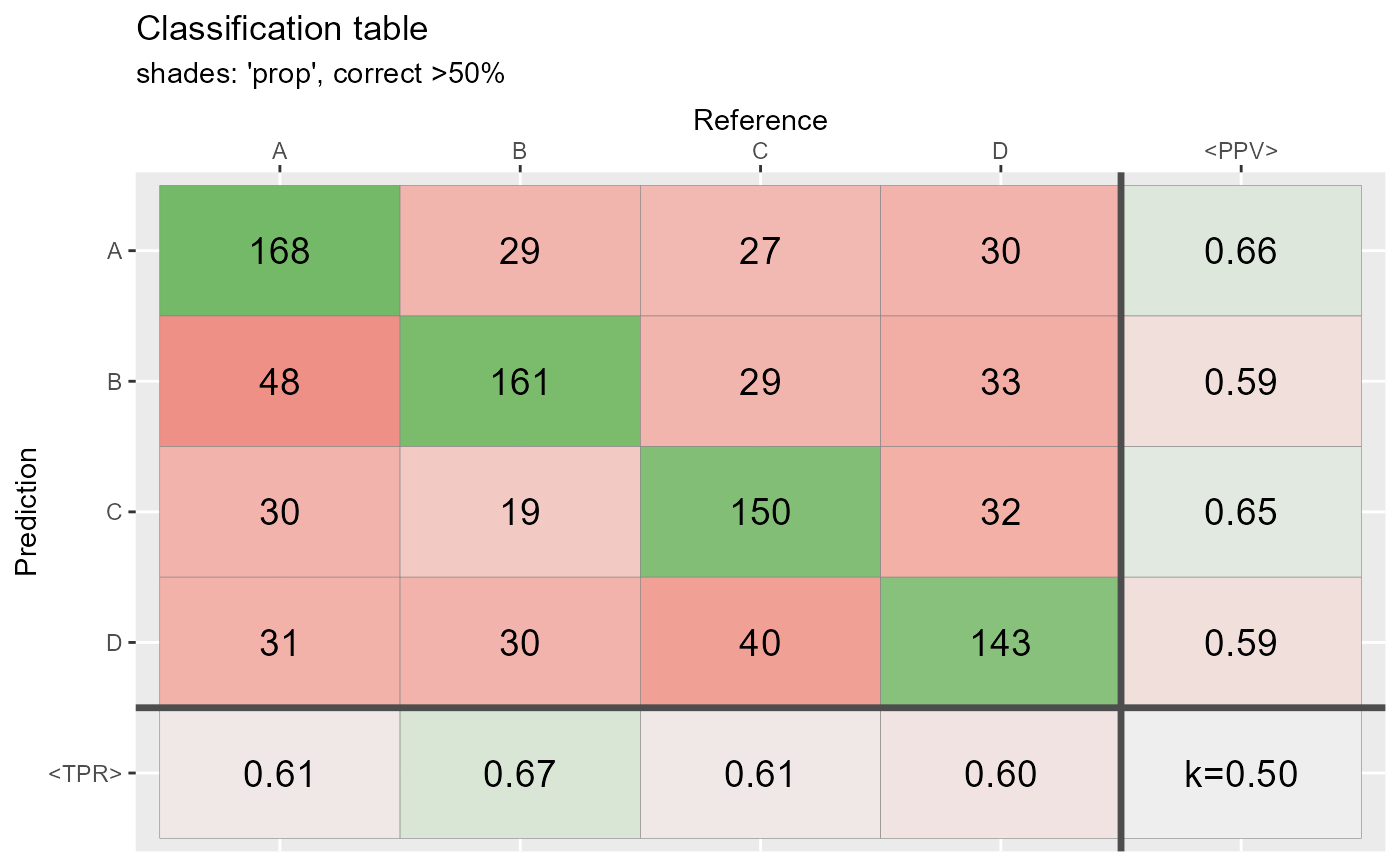 qplot_confusion(conf2,shades = "max", subTitle = "shades: 'max', correct >50%")
qplot_confusion(conf2,shades = "max", subTitle = "shades: 'max', correct >50%")
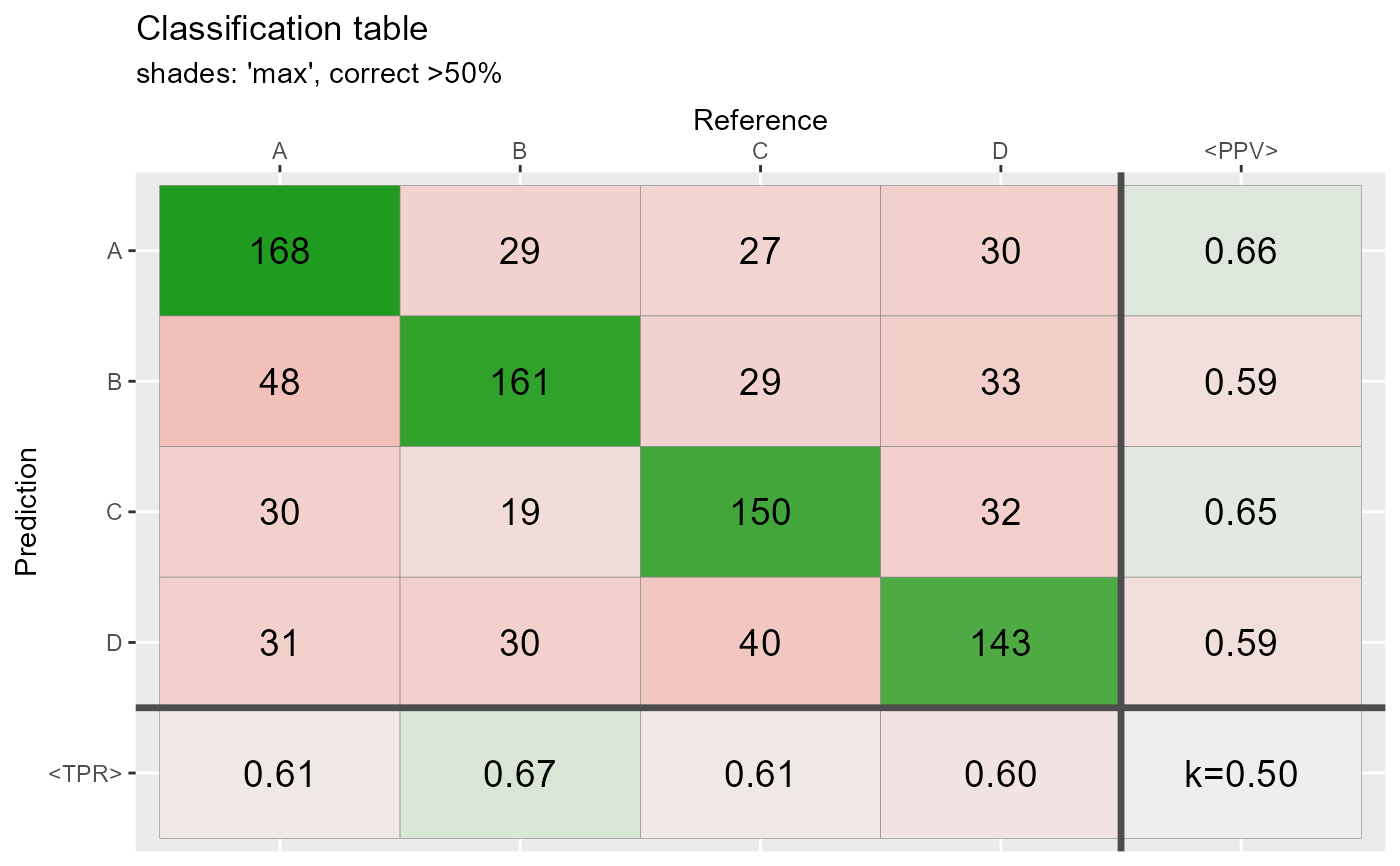 qplot_confusion(conf3,shades = "prop", subTitle = "shades: 'prop', correct >80%")
qplot_confusion(conf3,shades = "prop", subTitle = "shades: 'prop', correct >80%")
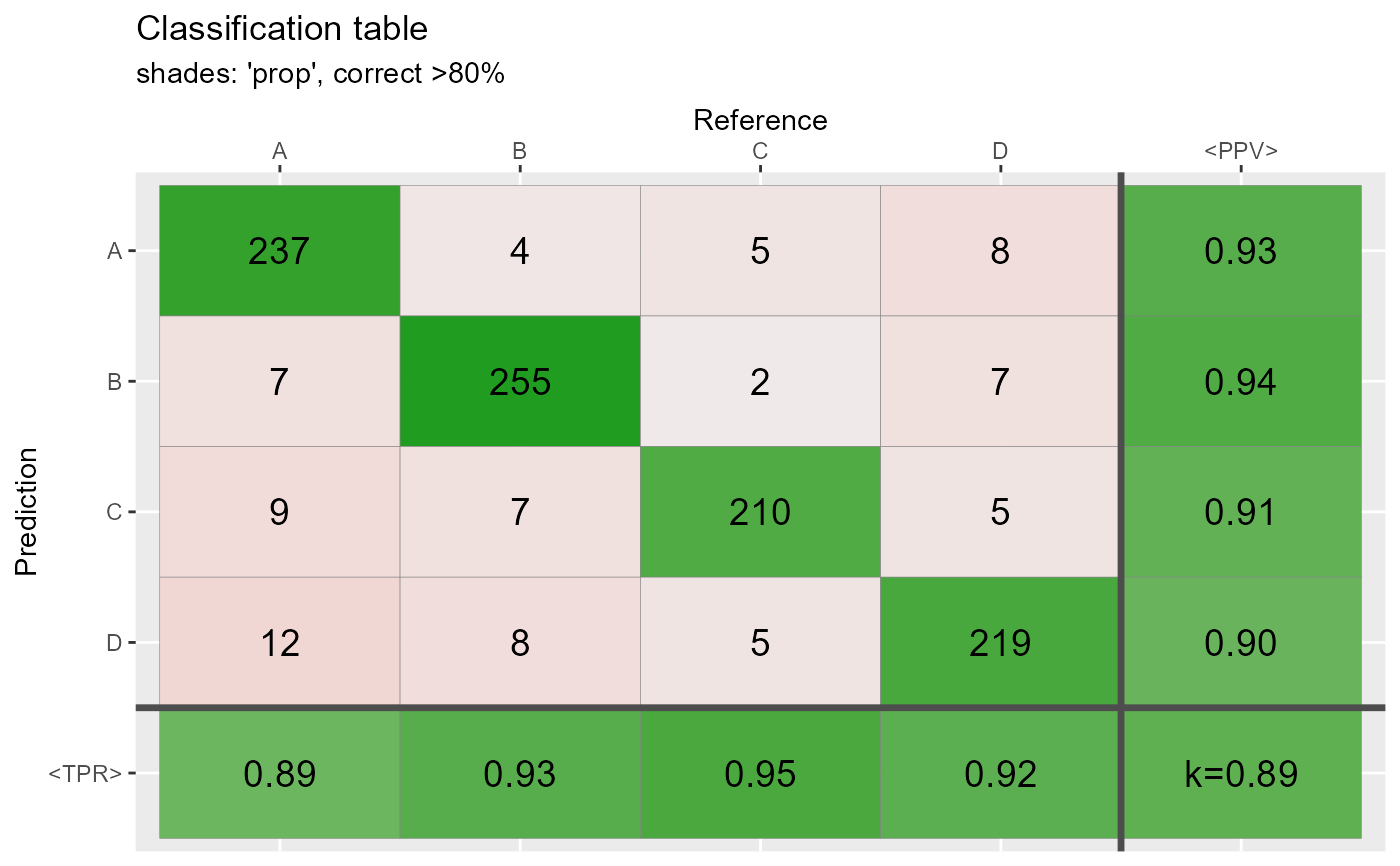 qplot_confusion(conf3,shades = "max", subTitle = "shades: 'max', correct >80%")
qplot_confusion(conf3,shades = "max", subTitle = "shades: 'max', correct >80%")
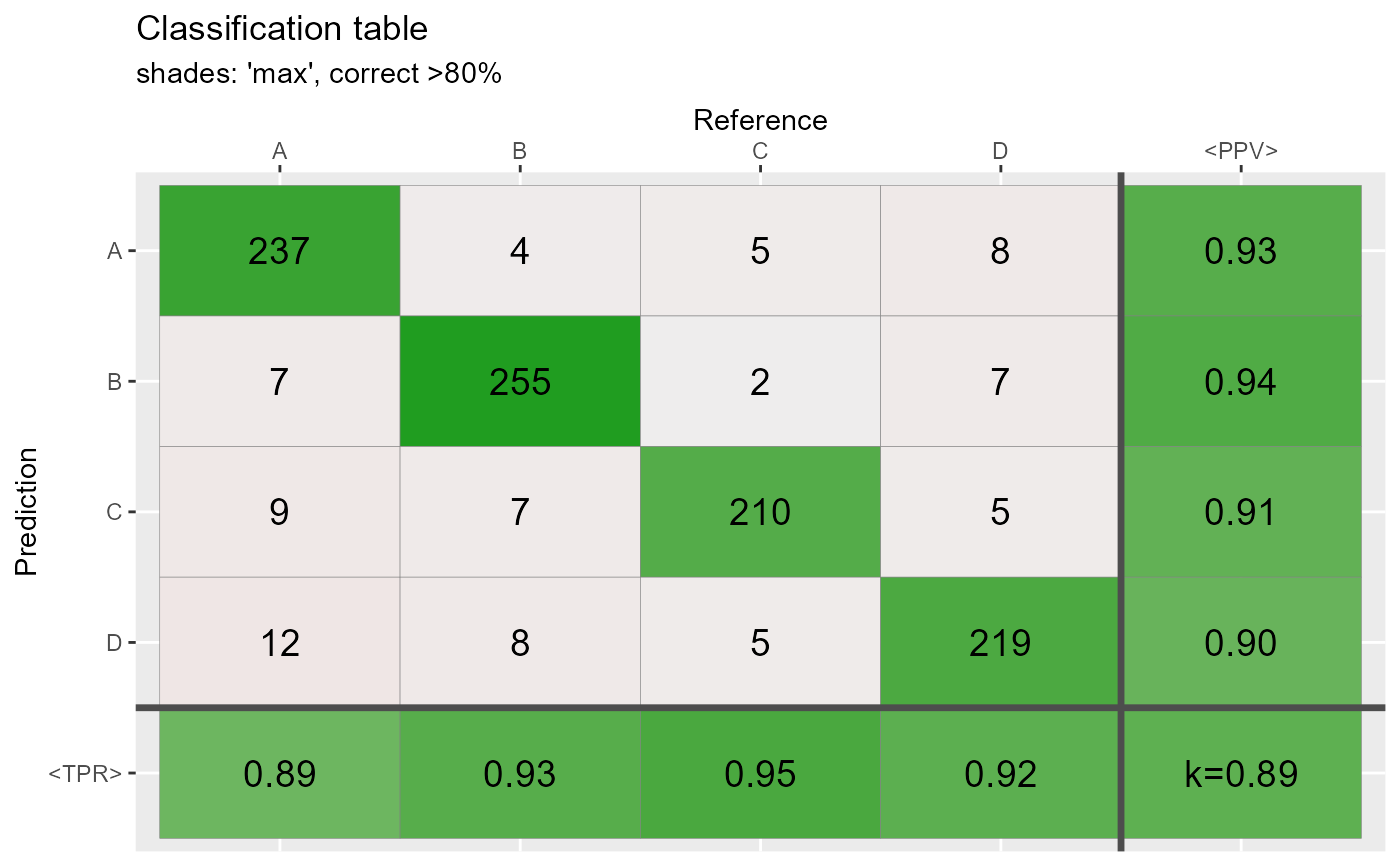 # Shades: constant and none ================================
qplot_confusion(conf3,shades = "const",subTitle = "shades: constant")
# Shades: constant and none ================================
qplot_confusion(conf3,shades = "const",subTitle = "shades: constant")
 qplot_confusion(conf3,shades = "none", subTitle = "shades: none")
qplot_confusion(conf3,shades = "none", subTitle = "shades: none")